近年来,免疫疗法彻底改变了癌症治疗。免疫疗法并非直接靶向肿瘤,而是引导患者的免疫系统更有效地攻击肿瘤。这对一些难以治疗的癌症尤其有效。然而,目前只有不到一半的癌症患者对免疫疗法有反应,因此迫切需要识别能够预测哪些患者最有可能从治疗中获益的生物标记物。
最近,科学家们注意到,肿瘤中 ARID1A 基因发生突变的患者更有可能对免疫检查点阻断产生积极反应,免疫疗法通过保持抗癌免疫细胞活性发挥作用。
由于 ARID1A 基因突变存在于许多癌症中,包括子宫内膜癌、卵巢癌、结直肠癌、胃癌、肝癌和胰腺癌,索尔克研究所的研究人员想知道它如何影响治疗敏感性,以及临床医生如何利用这些信息为每位患者提供个性化的癌症治疗。
他们发表在《细胞》杂志上的新研究表明,ARID1A 突变通过类似抗病毒的免疫反应将抗癌免疫细胞招募到肿瘤中,从而使肿瘤对免疫疗法敏感。
研究人员认为,这种突变和抗病毒免疫反应可以作为生物标志物,更好地筛选接受特定免疫疗法(例如免疫检查点阻断)的患者。这些发现也有助于开发针对ARID1A及其相关蛋白的药物,以提高其他肿瘤对免疫疗法的敏感性。
“这可能真正改变患者的癌症治疗结果,”该研究的资深作者、副教授戴安娜·哈格里夫斯说道。“携带ARID1A突变的患者已经产生了免疫反应,因此我们需要做的就是通过免疫检查点阻断来增强这种免疫反应,帮助他们从内部摧毁肿瘤。”
尽管已知携带ARID1A突变的人对免疫检查点阻断反应良好,但两者之间的确切联系仍不清楚。为了阐明这一机制,索尔克研究所的科学家使用了同时携带ARID1A突变和功能性ARID1A的黑色素瘤和结直肠癌小鼠模型。
来源:Cell (2024). DOI: 10.1016/j.cell.2024.04.025
研究小组在所有携带ARID1A突变的模型中都观察到了强烈的免疫反应,但在ARID1A功能正常的模型中却没有观察到,这支持了ARID1A突变确实驱动了这种反应的观点。但这在分子水平上是如何起作用的呢?
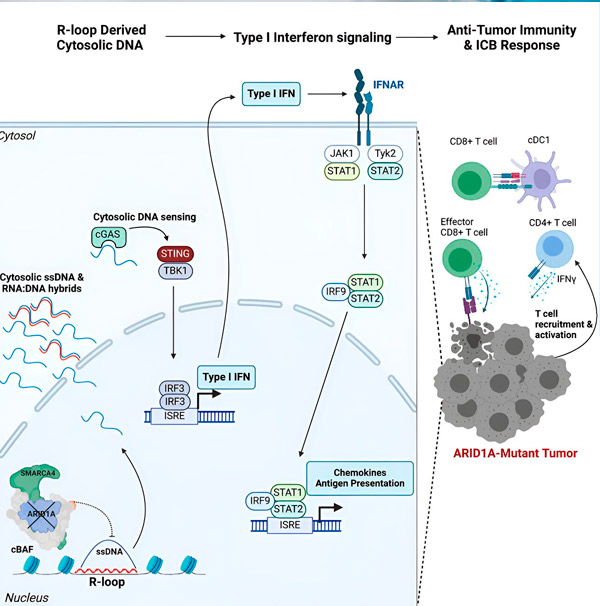
“我们发现ARID1A通过维持正确的DNA组织在细胞核中发挥着重要作用,”该研究的第一作者、Hargreaves实验室的研究生Matthew Maxwell说。“如果没有功能性的ARID1A,游离DNA就会被切除并释放到细胞溶胶中,从而激活所需的抗病毒免疫反应,而这种免疫反应可以通过免疫检查点阻断来增强。”
ARID1A基因编码一种蛋白质,它有助于调节DNA形状并维持基因组稳定性。当ARID1A发生突变时,它会在癌细胞中引发一系列类似鲁布·戈德堡机械的连锁反应。
首先,功能性ARID1A的缺失会导致DNA被释放到细胞质中。随后,细胞质DNA会激活抗病毒警报系统——cGAS-STING通路,因为我们的细胞适应于将细胞质中的任何DNA标记为外来物质,以防止病毒感染。最终,cGAS-STING通路会启动免疫系统,将T细胞募集到肿瘤部位,并将其激活为专门的癌症杀伤T细胞。
ARID1A突变、DNA逃逸、cGAS-STING警报、T细胞募集等一系列事件,每一步都依赖于前一步,导致肿瘤中抗癌T细胞的数量增加。免疫检查点阻断可以用来确保这些T细胞保持活性,从而增强其抗癌能力。
“我们的研究结果提供了一种新的分子机制,ARID1A 突变可能通过该机制促进抗肿瘤免疫反应,”Hargreaves 说道。“这些发现令人兴奋的地方在于它们的转化潜力。我们可以利用 ARID1A 突变来筛选接受免疫检查点阻断的患者,现在我们发现了一种机制,即抑制 ARID1A 或其蛋白质复合物的药物可以用来进一步增强其他患者的免疫疗法。”
通过描述免疫检查点阻断在ARID1A突变癌症中更有效的机制,研究人员为临床医生提供了优先为ARID1A突变患者提供免疫疗法的理论依据。这些发现是迈向个性化癌症治疗的重要一步,并启发了针对ARID1A及其蛋白质复合物的新疗法的开发。
展望未来,索尔克研究所团队希望他们的研究结果能够改善与 ARID1A 突变相关的各种癌症患者的治疗结果,并打算与加州大学圣地亚哥分校合作探索这一临床转化。

